Check out our new app note with @Diagenode! Best practices when using #Megaruptor3 w/ #PippinHT or #BluePippin. Includes our new Range+T size selection method & tips on getting tight HMW DNA in the 9-30kb range. #PacBio #HiFisequencing #nanopore #genomics https://sagescience.com/blog/ranget-for-tight-sizing-of-hmw-libraries/
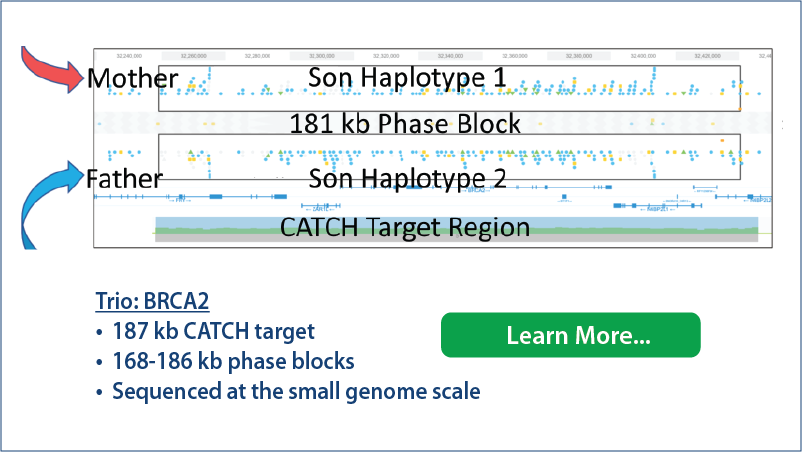
Phased genomic targets from @universal_seq . Very impressive paper! #TELLseq -- linked reads in a tube. Check it out!
#longreads #HLSCATCH #phasedgenomes #NGS #illumina #linkedreads
Long reads are getting longer – again! | Arizona Genomics Institute
@PacBio @dariocopet @SageSci